
Get started with browseMetadata
browseMetadata.RmdFor installation, set-up and basic usage refer to the package README.md file.
This page provides more context on how to interact with package functions and interpret package outputs.
browseMetadata()
The json file contains information about the data asset, data class and each data element. In the metadata catalogue:
- Data asset refers to a Dataset (a collection of data, can contain multiple tables)
- Data class refers to a Table within a dataset
- Data Element refers to each Variable within a table
See here for outputs generated from a demo run.
- BROWSE_table html summarises the dataset and each table in the dataset
- BROWSE_bar html, pasted below, is a simple bar plot summarising the dataset
- BROWSE_bar csv file contains the data used to make this bar plot
_V16.0.0.png)
We can see there are 13 tables in the dataset. The (numbers) next to
table names correspond to the order in which they are shown to you in
the mapMetadata() function. The height of the bar indicates
the number of variables in that table:
- The ones with lots of variables (e.g. CHILD_TRUST) will take you
longer to process when running
mapMetadata() - Some tables (e.g. CHE_HEALTHYCHILDWALESPROGRAMME) have a lot of empty descriptions. An empty description means that this variable will only have a label and a data type.
It is important to note that this plot is only summarising variable level metadata i.e. a description of what the variable is. Some variables also require value level metadata i.e. what does each value correspond to, 1 = Yes, 2 = No, 3 = Unknown. This value level metadata can sometimes be found in lookup tables, if it is not provided within the variable level description.
mapMetadata()
Running the function in demo mode will use the same demo json file as
browseMetadata():
Demo mode only processes the first 20 variables (data elements) within the table(s) we select to process.
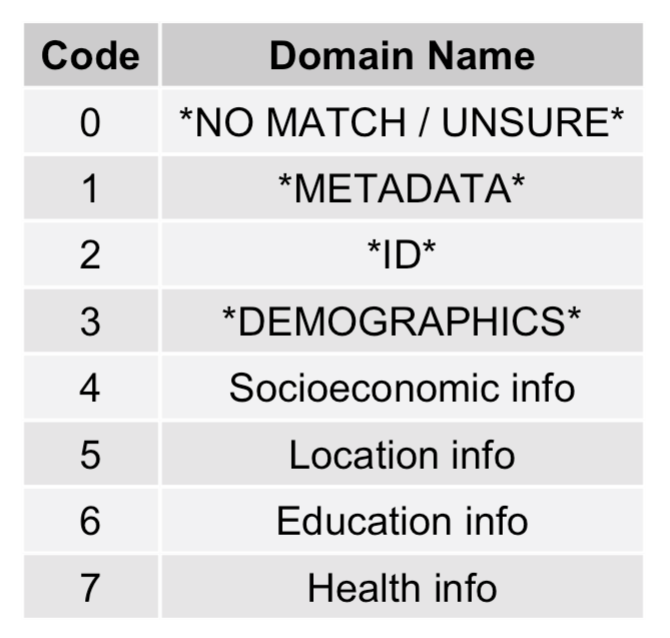
You will be asked to label data elements with one (or more) of the numbers shown in the Plots tab [0-7]. Here we have very simple domains [4-7] for the demo run. For a research study, your domains are likely to be much more specific e.g. ‘Prenatal, antenatal, neonatal and birth’ or ‘Health behaviours and diet’. The 4 default domains are always included [0-3], appended on to any domain list given.
ℹ Running mapMetadata in demo mode using package data files
ℹ Using the default look-up table in data/look-up.rda
Enter your initials: RSRespond with your initials after the prompt and press enter. It will then print the name of the dataset and where it was retrieved from:
── Dataset Name ─────────────────────────────────────────────────────────────────────────────────────────────────
National Community Child Health Database (NCCHD)
── Dataset File Exported By ─────────────────────────────────────────────────────────────────────────────────────
Rachael Stickland at 2024-04-05T13:01:23.109Z
ℹ Reference outputs from browseMetadata for information about the dataset
Press any key to continue Table_Name Table_Number
EXAM 1
CHILD 2
REFR_IMM_VAC 3
IMM 4
BREAST_FEEDING 5
PATH_BLOOD_TESTS 6
CHE_HEALTHYCHILDWALESPROGRAMME 7
BLOOD_TEST 8
CHILD_TRUST 9
PATH_SPCM_DETAIL 10
CHILD_MEASUREMENT_PROGRAM 11
CHILD_BIRTHS 12
SIG_COND 13
ℹ Found 13 table(s) in this Dataset. Enter table numbers you want to process (one table number on each line):
1: 2
2: For the purpose of this demo, type 2 to just process the CHILD table only. Leave the prompt on the second row blank and press enter.
To process multiple tables at once in the same session (e.g. CHILD, SIG_COND) include their numbers on multiple lines:
ℹ Enter each table number you want to process in this interactive session.
1: 1
2: 13
3:ℹ Processing Table 2 of 13
── Table Name ───────────────────────────────────────────────────────────────────────────────────────────────────
CHILD
ℹ Reference outputs from browseMetadata for information about the table
Optional free text note about this table (or press enter to continue): This table is important because ... It will now start looping through the data elements. If it skips over one it means it was auto-categorised or copied from a previous table already processed (more on that later). For this demo, it will only process 20 data elements (out of the 35 total).
ℹ 20 left to process in this session
✔ Processing data element 1 of 35
ℹ 19 left to process in this session
✔ Processing data element 2 of 35
ℹ 18 left to process in this session
✔ Processing data element 3 of 35
ℹ 17 left to process in this session
✔ Processing data element 4 of 35
DATA ELEMENT -----> APGAR_1
DESCRIPTION -----> APGAR 1 score. This is a measure of a baby's physical state at birth with particular reference to asphyxia - taken at 1 minute. Scores 3 and below are generally regarded as critically low; 4-6 fairly low, and 7-10 generally normal. Field can contain high amount of unknowns/non-entries.
DATA TYPE -----> CHARACTER
Categorise data element into domain(s). E.g. 3 or 3,4: 7
Categorisation note (or press enter to continue): your note here We chose to respond with ‘7’ because that corresponds to the ‘Health info’ domain in the table. More than one domain can be chosen. Do remember that this demo has over-simplified domain labels, and they will likely be more specific for a research study.
You have the option to re-do the categorisation (and note) you just made, by replying ‘y’ to the question:
Response to be saved is '7'. Would you like to re-do? (y/n): yAfter completing 20, it will then ask you to review the auto-categorisations it made.
These auto-categorisations are based on the mappings included in the
default look_up.csv.
Type get("look_up") in R.
This look-up file can be changed by the user. ALF refers to ‘Anonymous Linking Field’ - this field is used within datasets that have been anonymised and encrypted for inclusion within SAIL Databank.
DataElement Domain_code Note
1 ALF_E 2 AUTO CATEGORISED
2 ALF_MTCH_PCT 2 AUTO CATEGORISED
3 ALF_STS_CD 2 AUTO CATEGORISED
6 AVAIL_FROM_DT 1 AUTO CATEGORISED
19 GNDR_CD 3 AUTO CATEGORISED
ℹ These are the auto categorised data elements. Enter row numbers for those you want to edit:
1: Press enter for now. It will then ask you if you want to review the categorisations you made. Respond Y to review:
Would you like to review your categorisations? (y/n): y
DataElement Domain_code Note (first 12 chars)
4 APGAR_1 7
5 APGAR_2 7
7 BIRTH_ORDER 7 10% missingness
8 BIRTH_TM 1,7 20% missingness
9 BIRTH_WEIGHT 7
10 BIRTH_WEIGHT_DEC 7
11 BREASTFEED_8_WKS_FLG 7
12 BREASTFEED_BIRTH_FLG 7
13 CHILD_ID_E 2
14 CURR_LHB_CD_BIRTH 5,7 Place of birth
15 DEL_CD 7
16 DOD 3,7
17 ETHNIC_GRP_CD 3
18 GEST_AGE 3,7
20 HEALTH_VISITOR_CD_E 2
ℹ Press enter to accept your categorisations for table CHILD, or enter each row number you'd like to edit:
1: 8
2: 14
3: If you want to change your categorisation, enter in the row number (e.g. 8 for BIRTH_TM and 14 for CURR_LHB_CD_BIRTH).
It will then take you through the same process as before, and you can over-write your previous categorisation.
All finished! Take a look at the outputs:
✔ Your final categorisations have been saved:
OUTPUT_NationalCommunityChildHealthDatabase(NCCHD)_CHILD_2024-04-05-14-37-36.csv
✔ Your session log has been saved:
LOG_NationalCommunityChildHealthDatabase(NCCHD)_CHILD_2024-04-05-14-37-36.csv
✔ A summary plot has been saved:
PLOT_NationalCommunityChildHealthDatabase(NCCHD)_CHILD_2024-04-05-14-37-36.pngThe OUTPUT csv contains the categorisations you made. The LOG csv
contains information about the session as a whole, including various
metadata. These two csv files contain the same timestamp column. If you
do not like the formatting of the OUTPUT csv, see the function
?mapMetadata_convert_outputs for an alternative.
The PLOT png file saves a simple plot displaying the count of domain codes for that table.